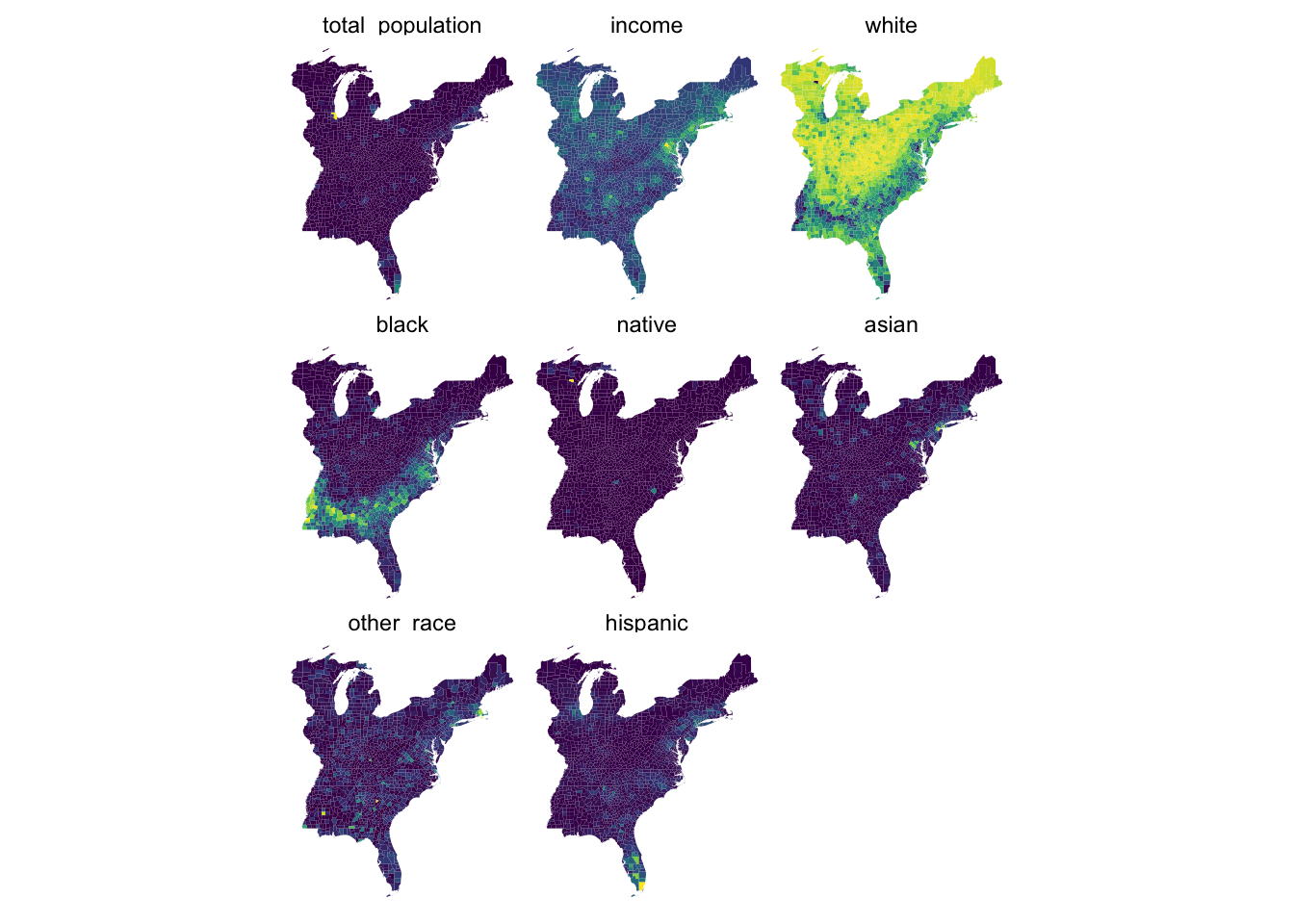
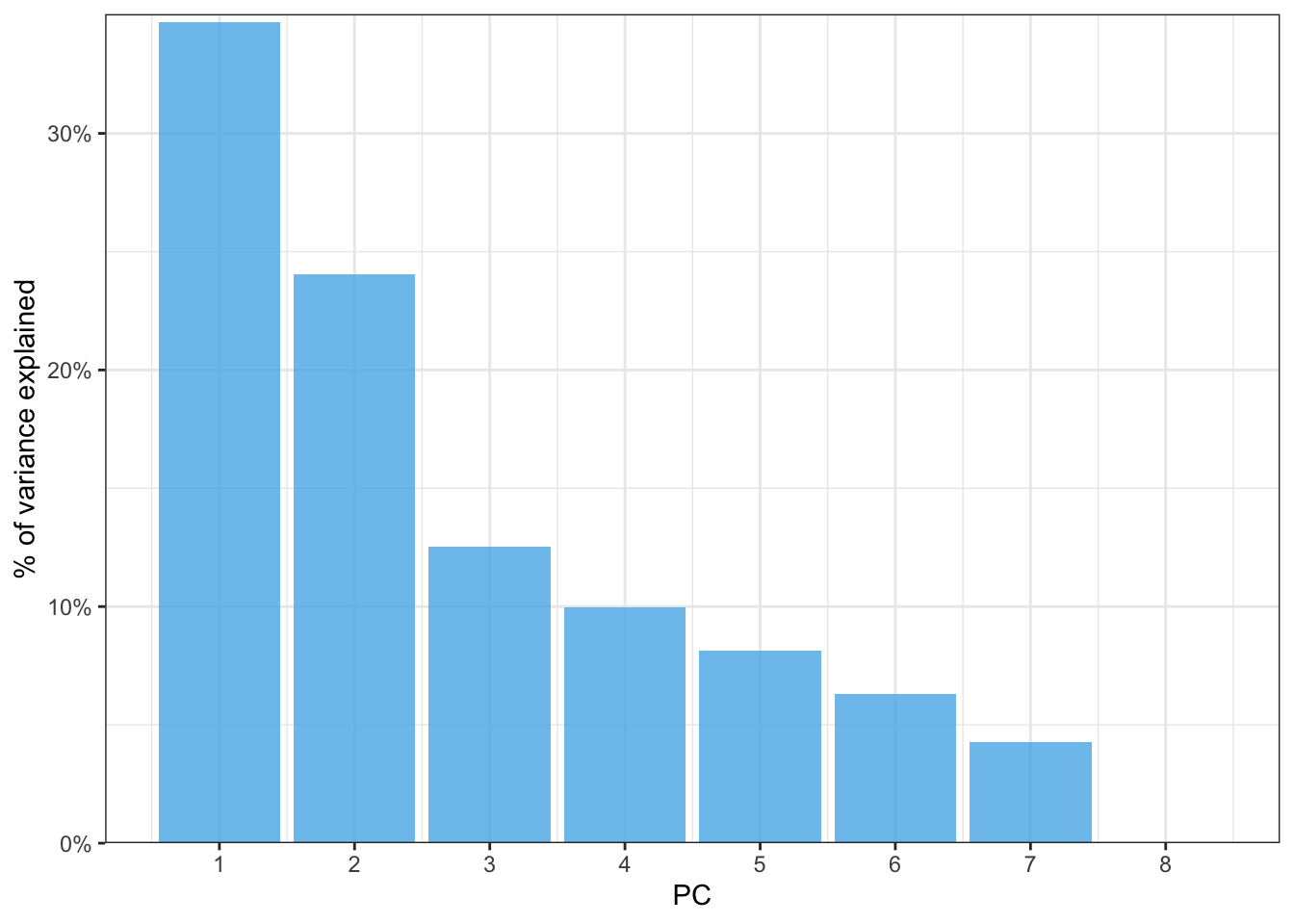
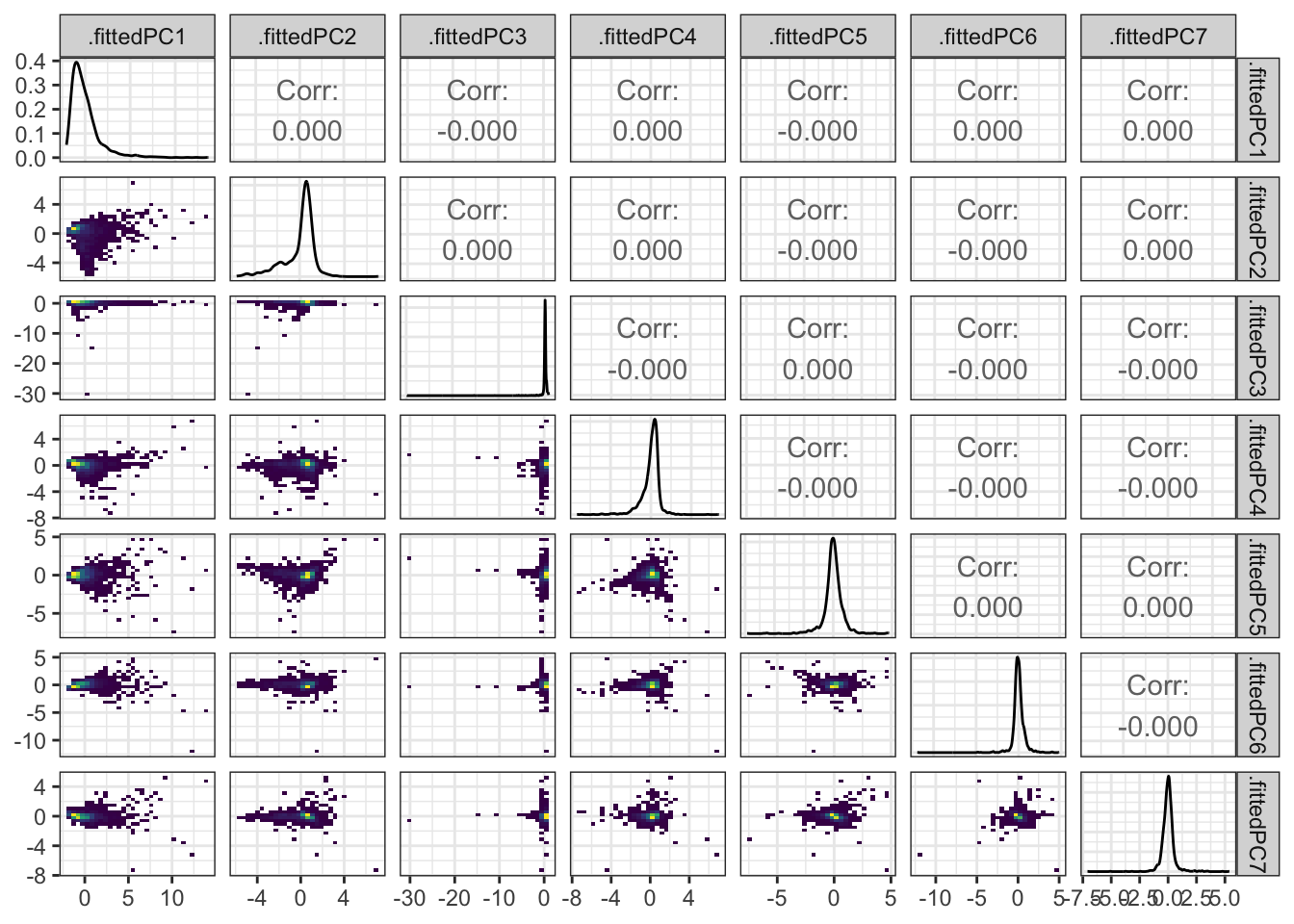
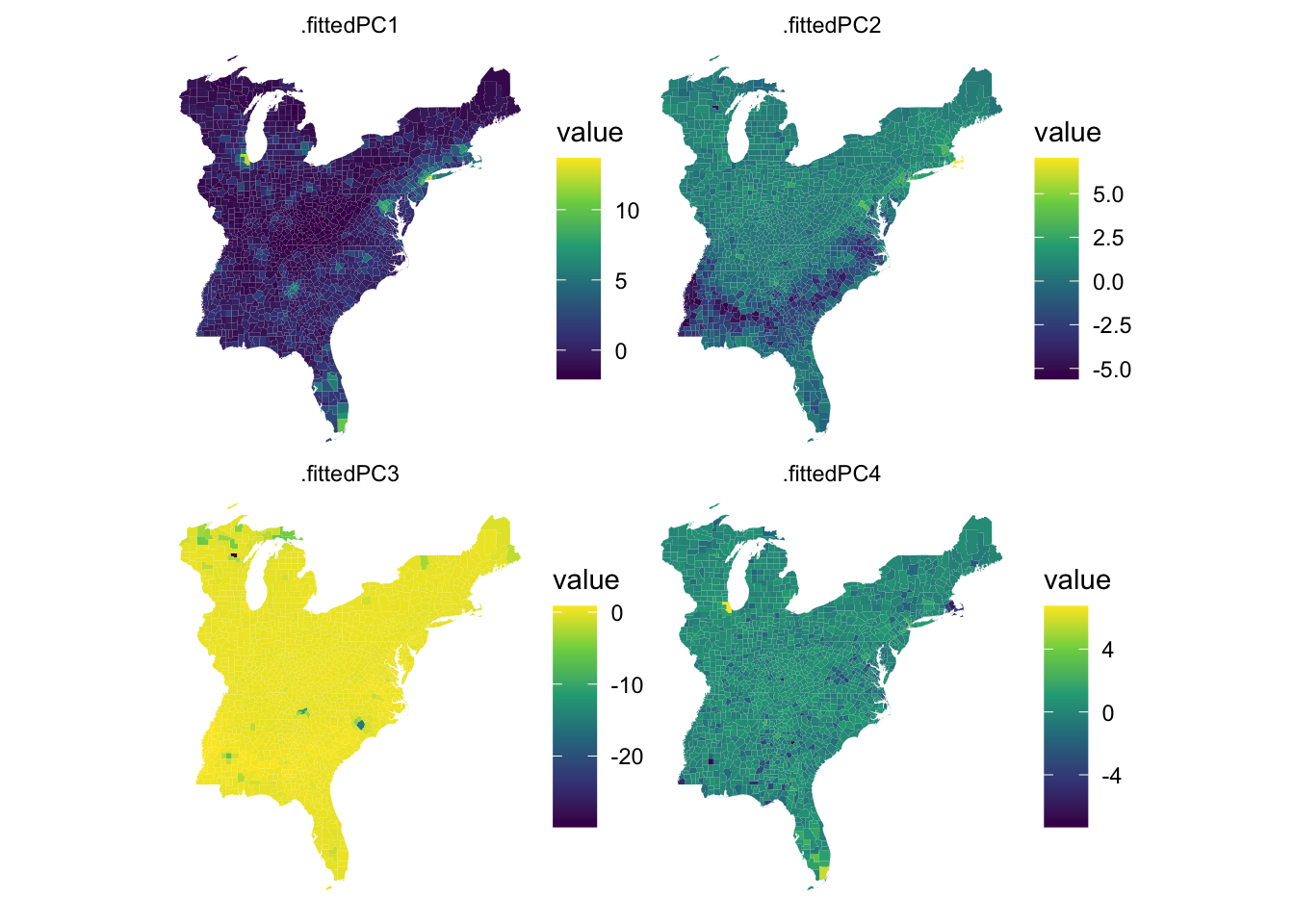
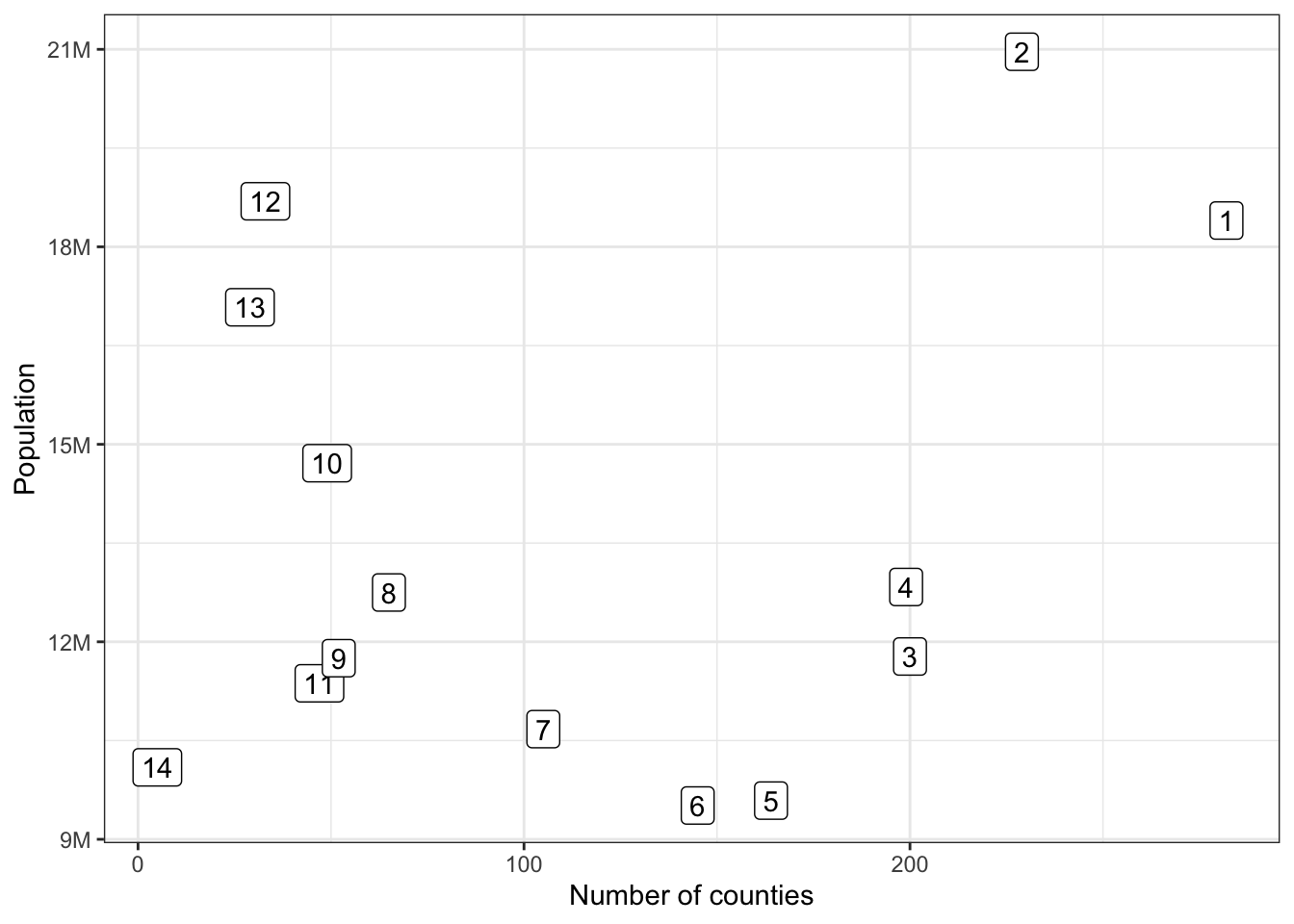
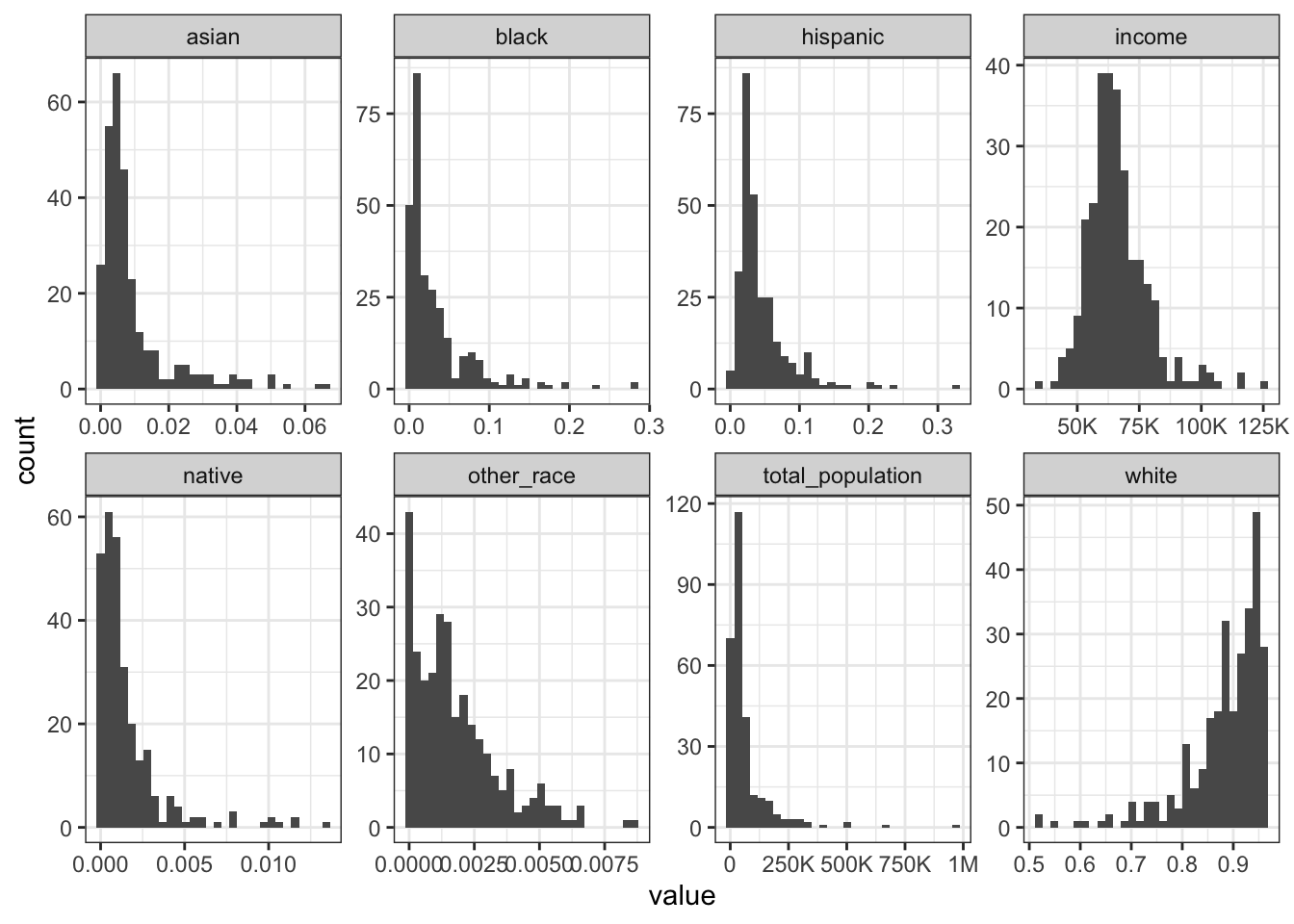
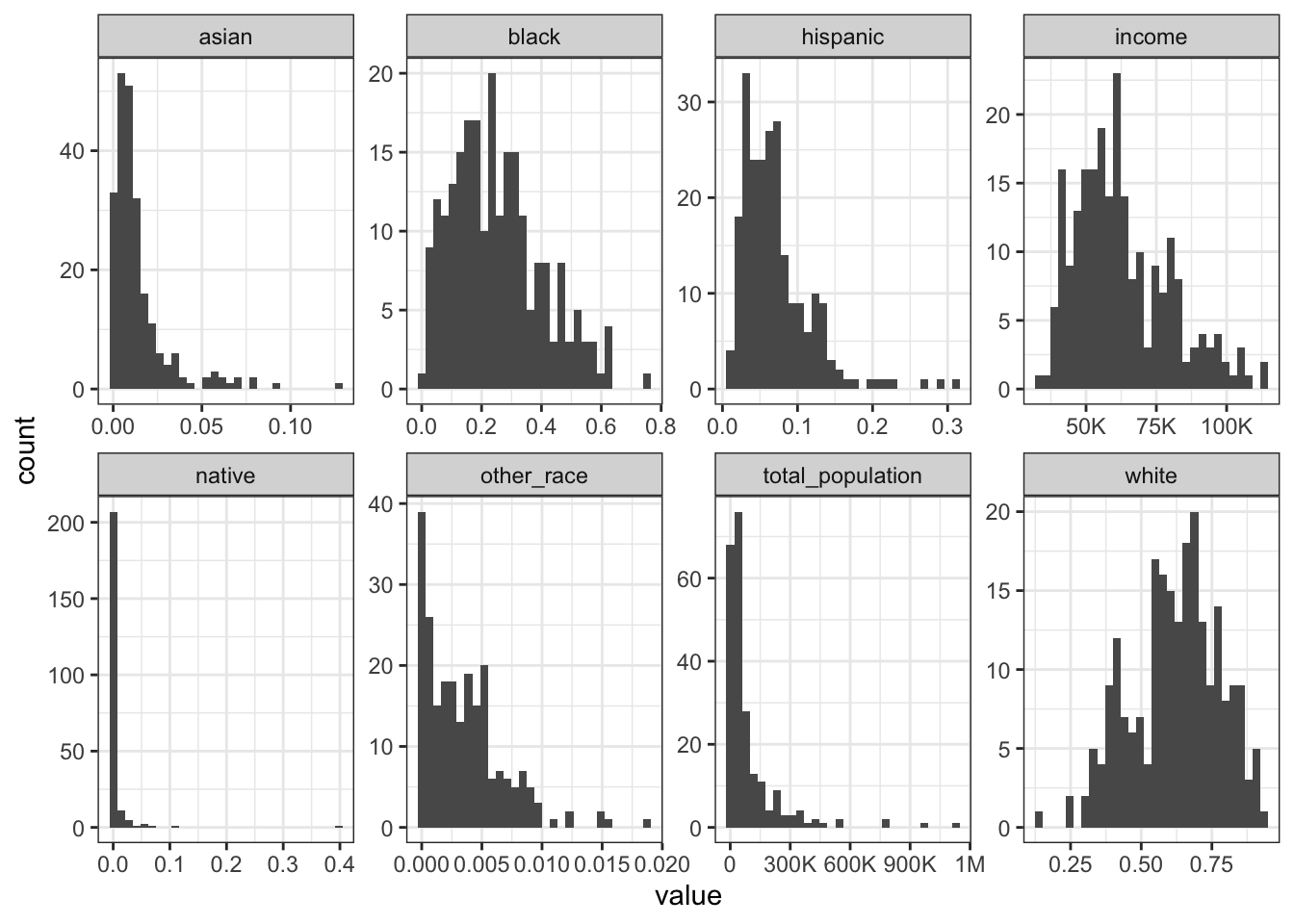
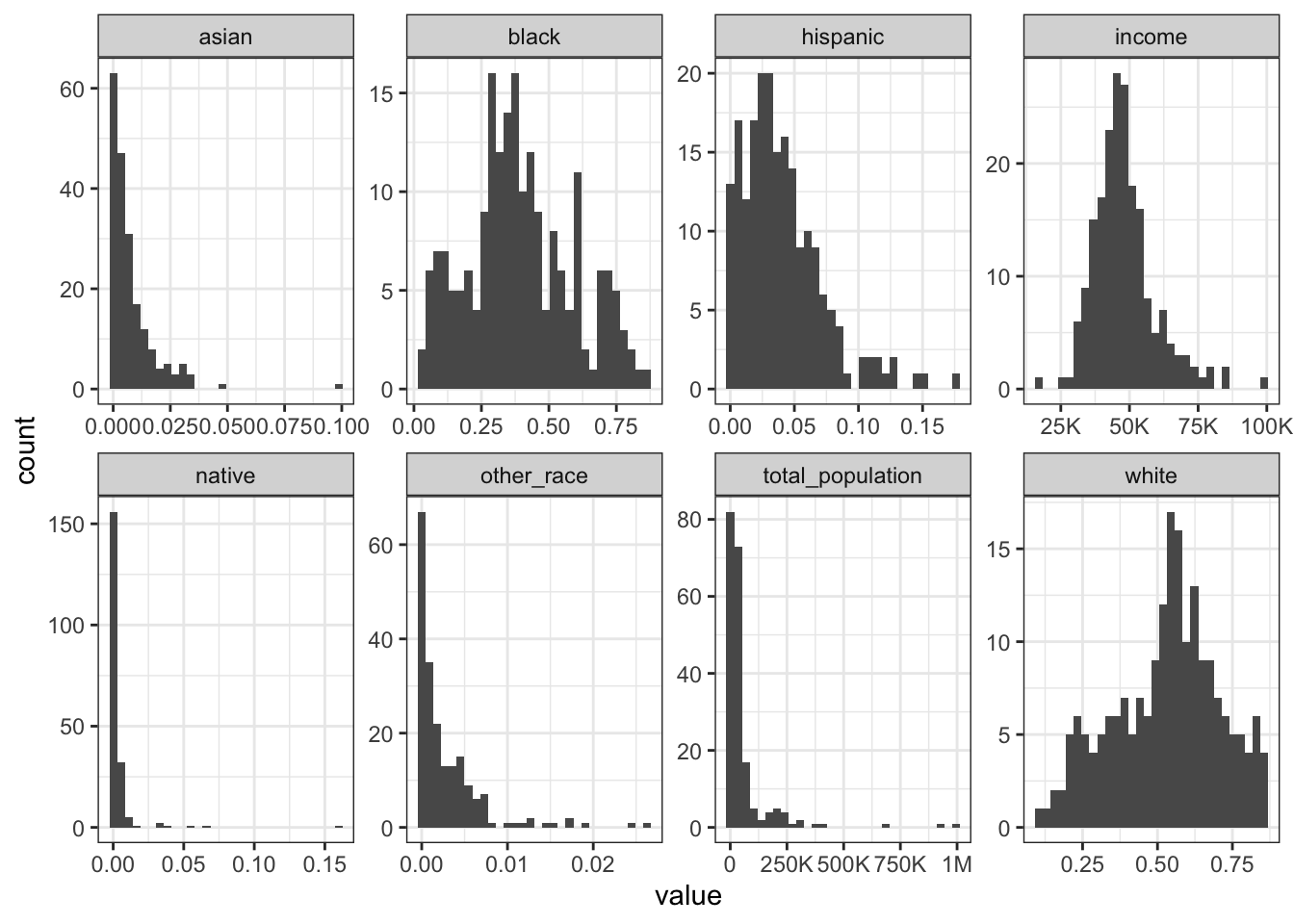
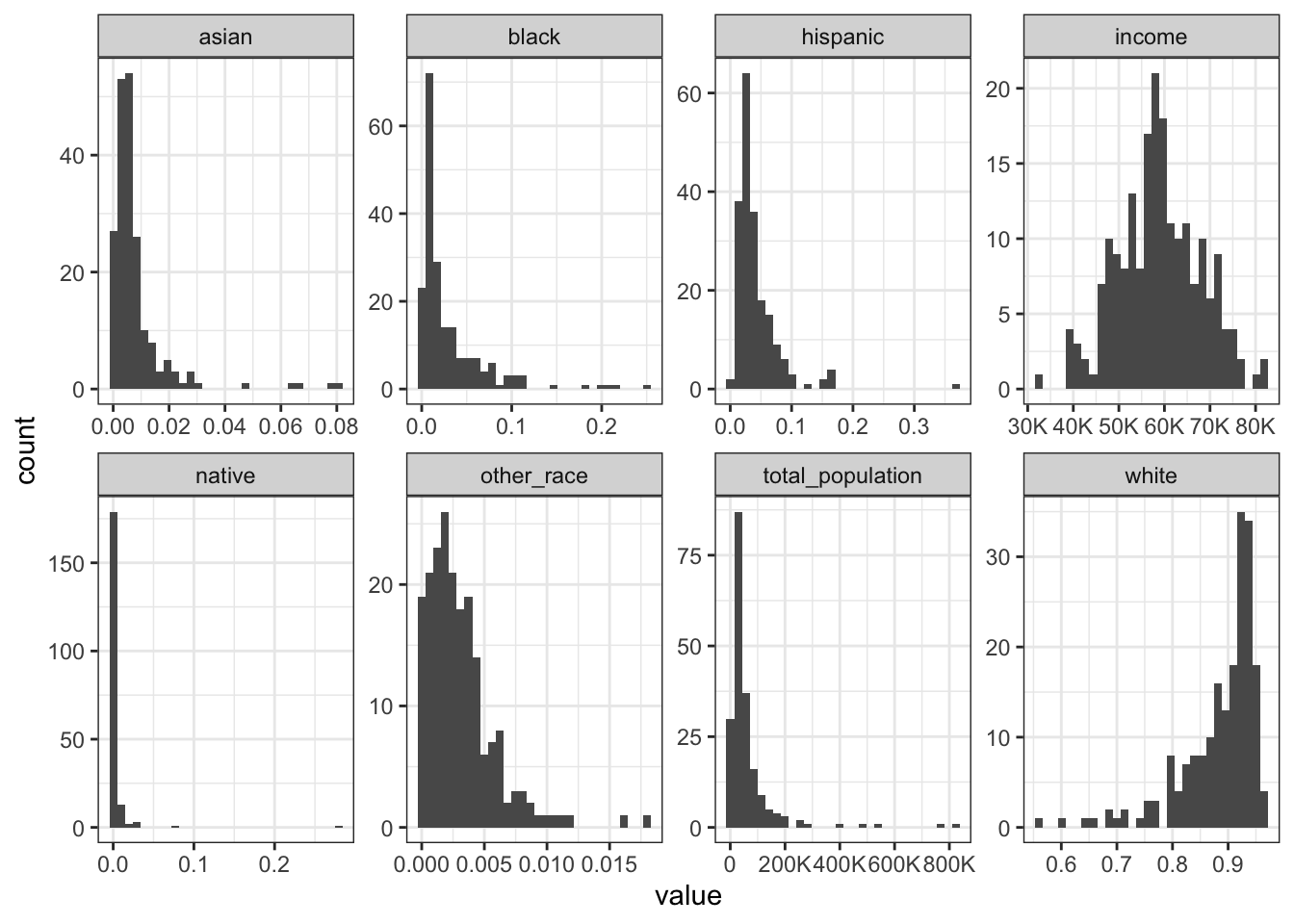
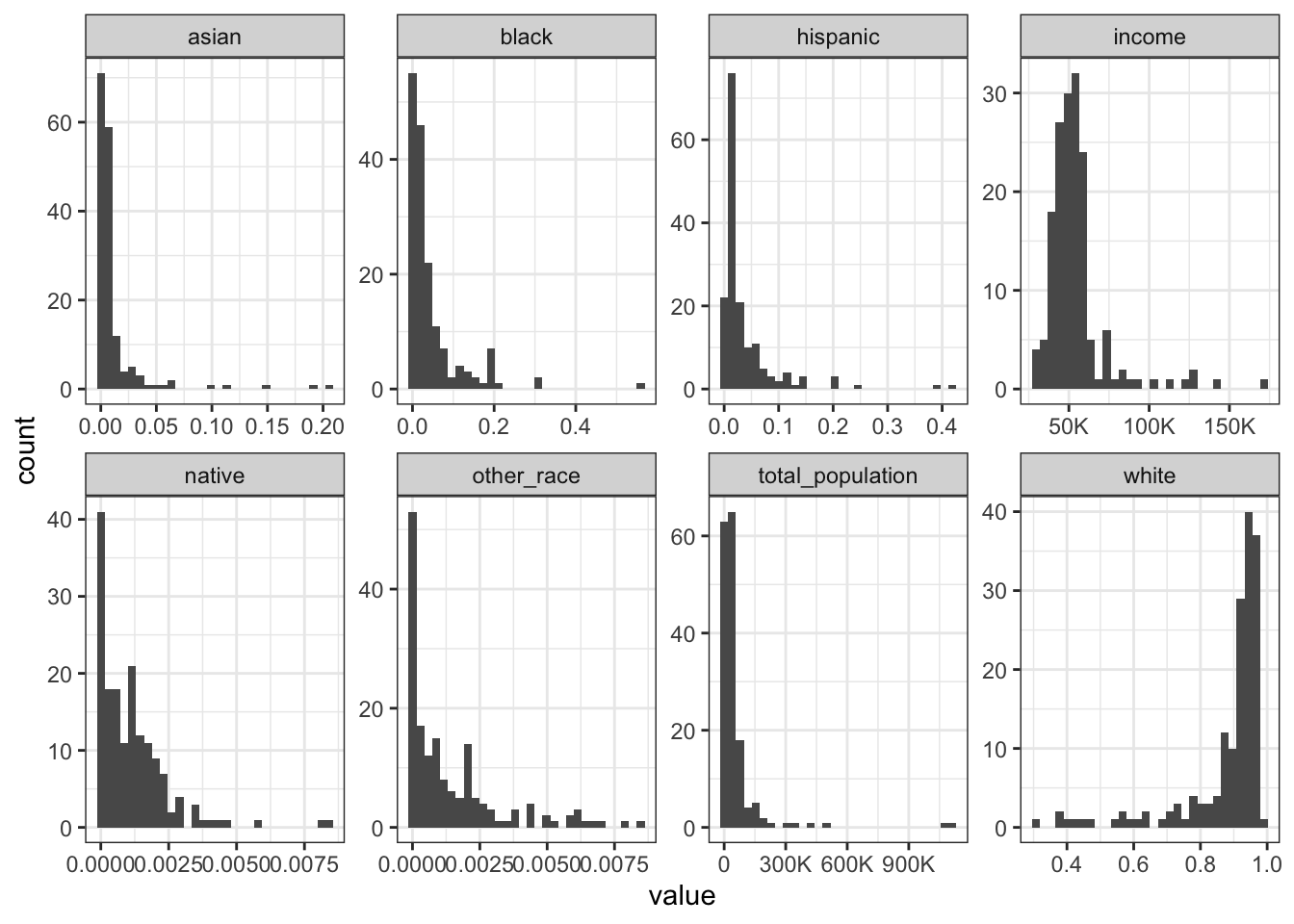
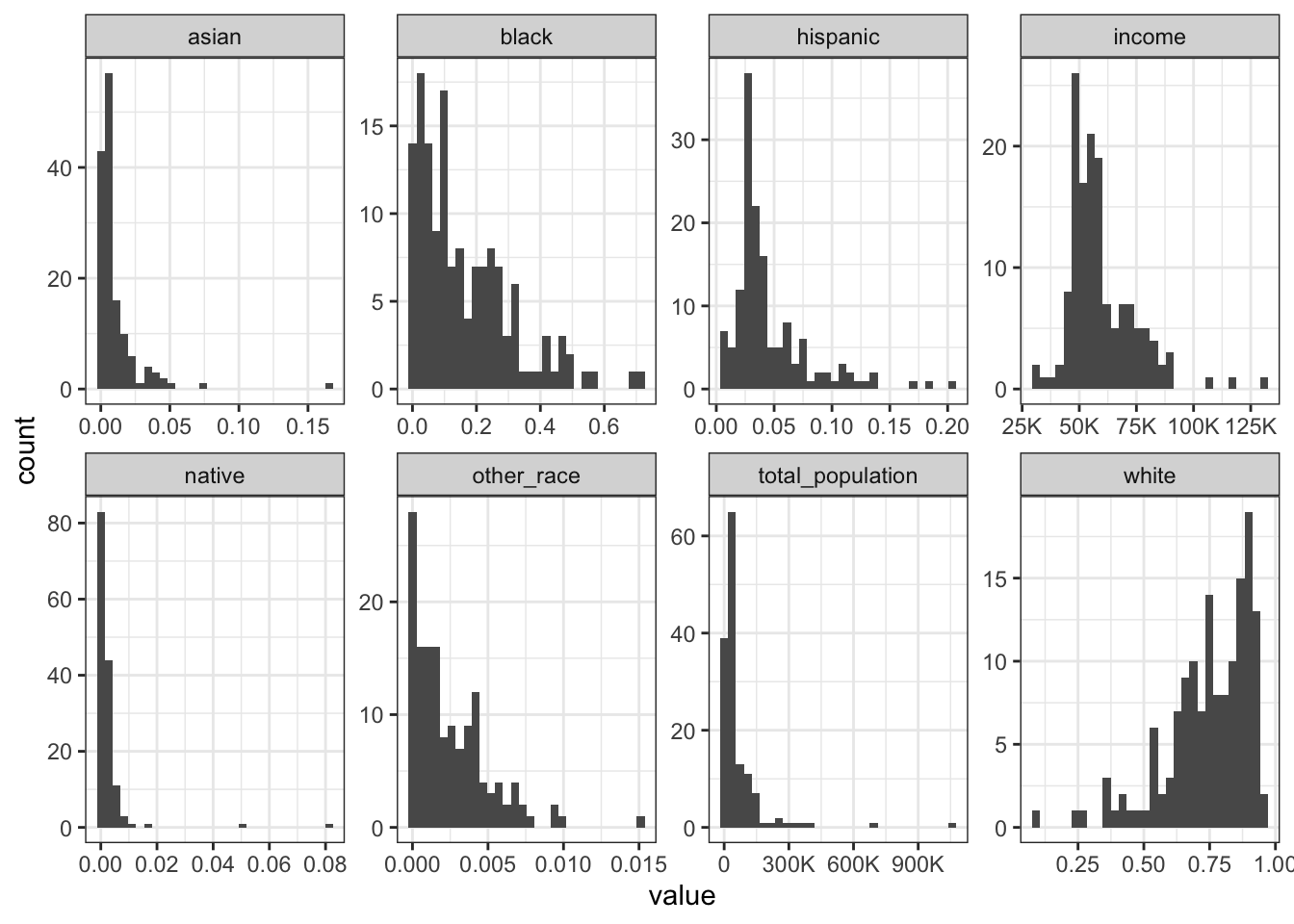
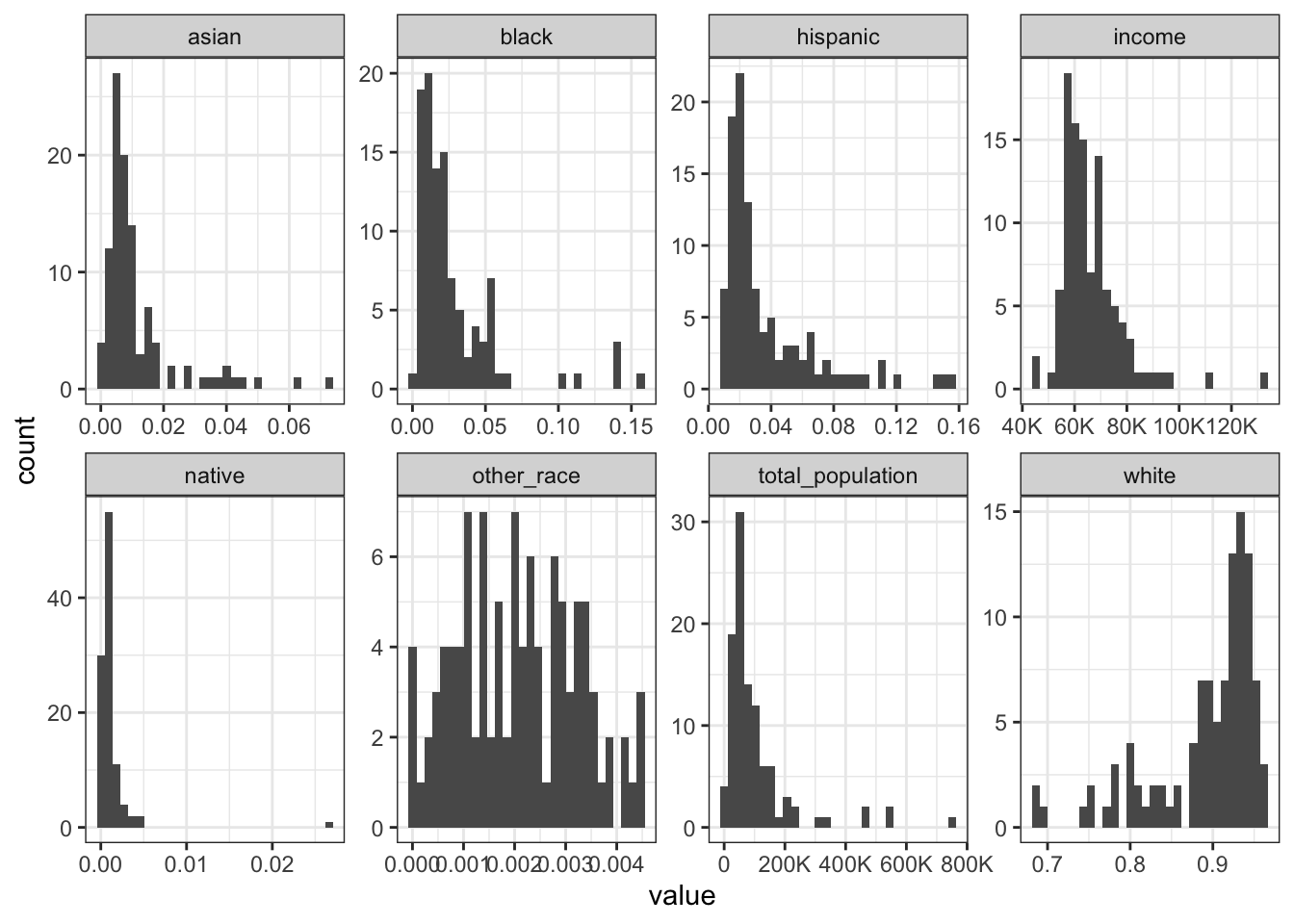
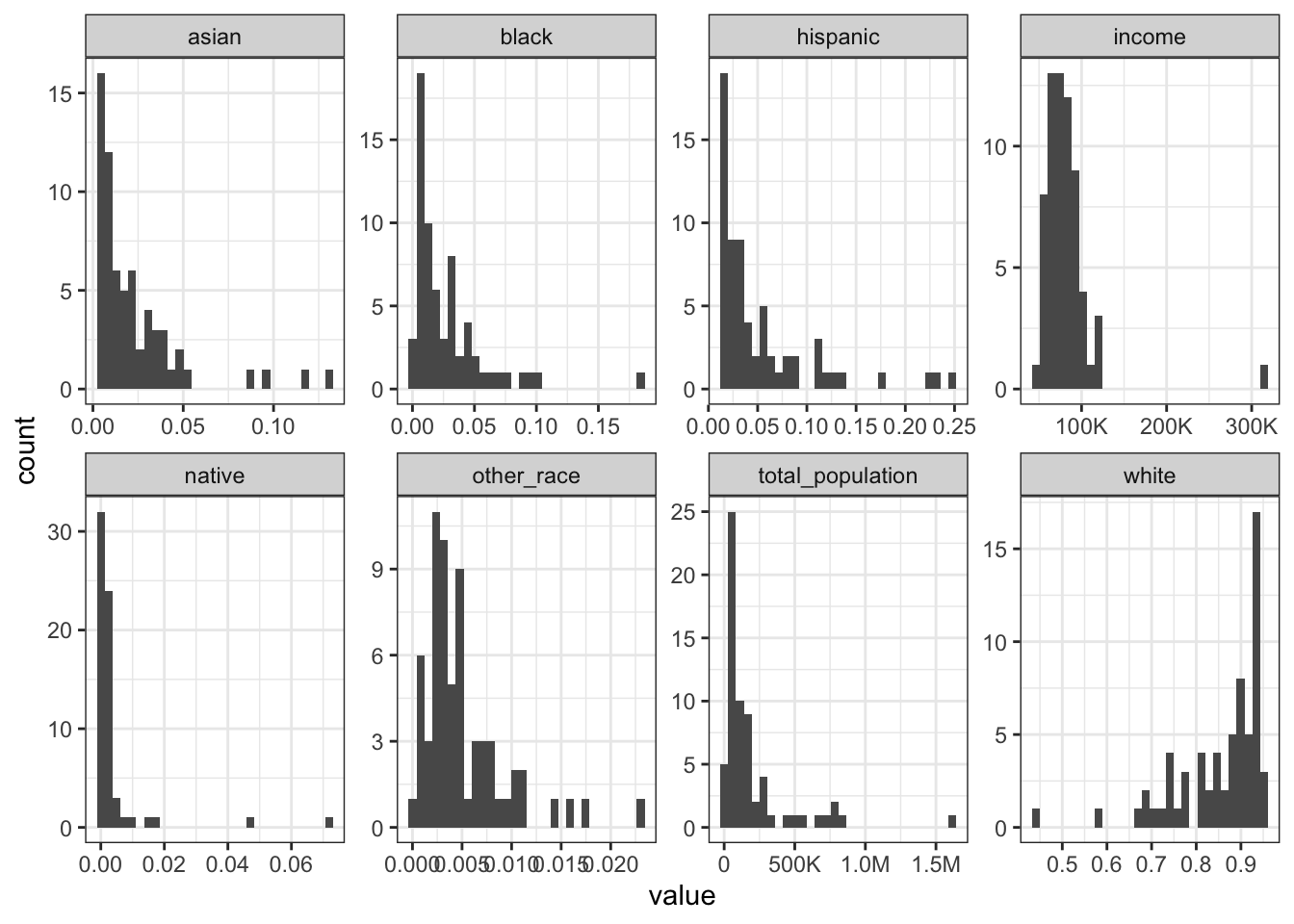
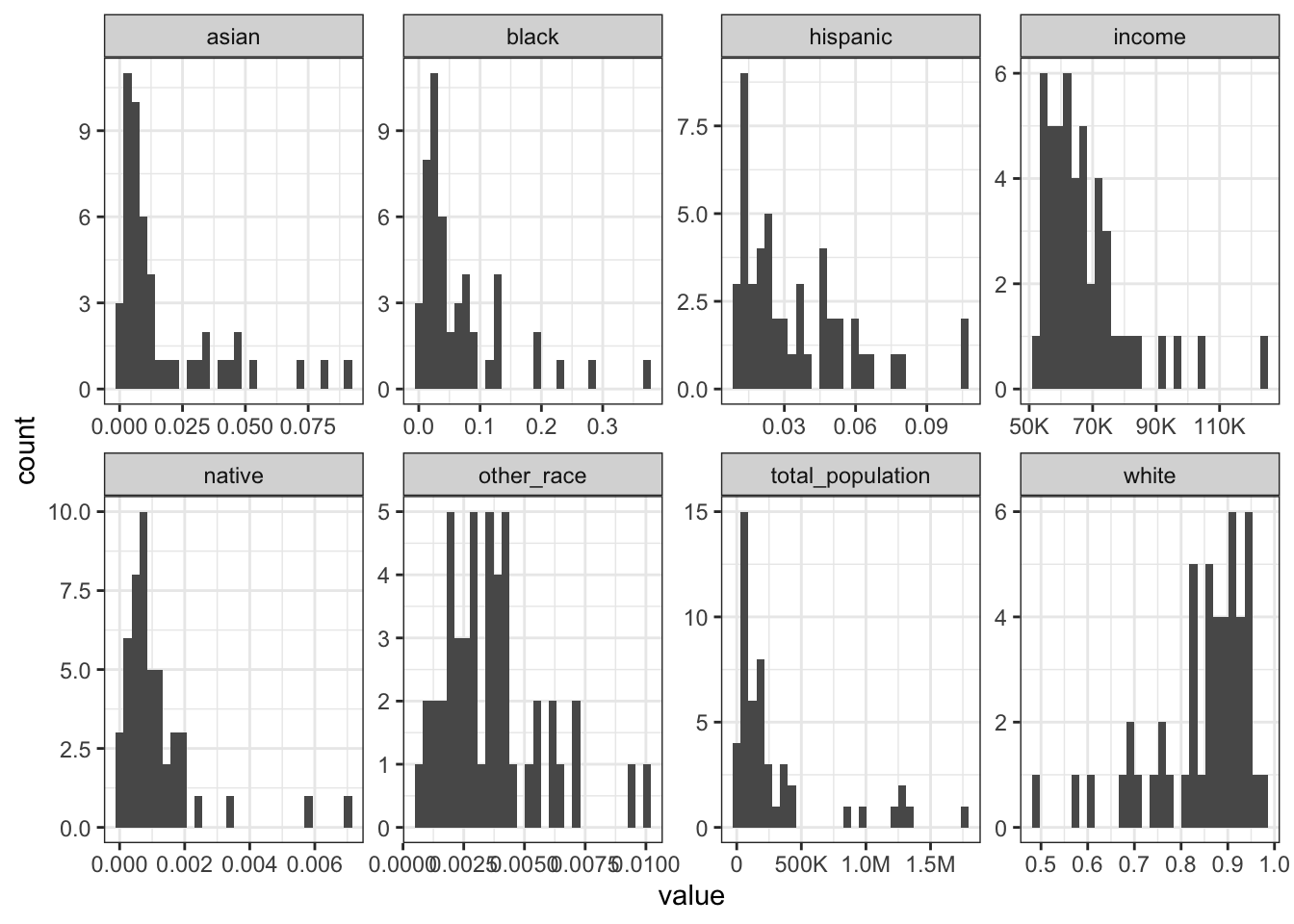
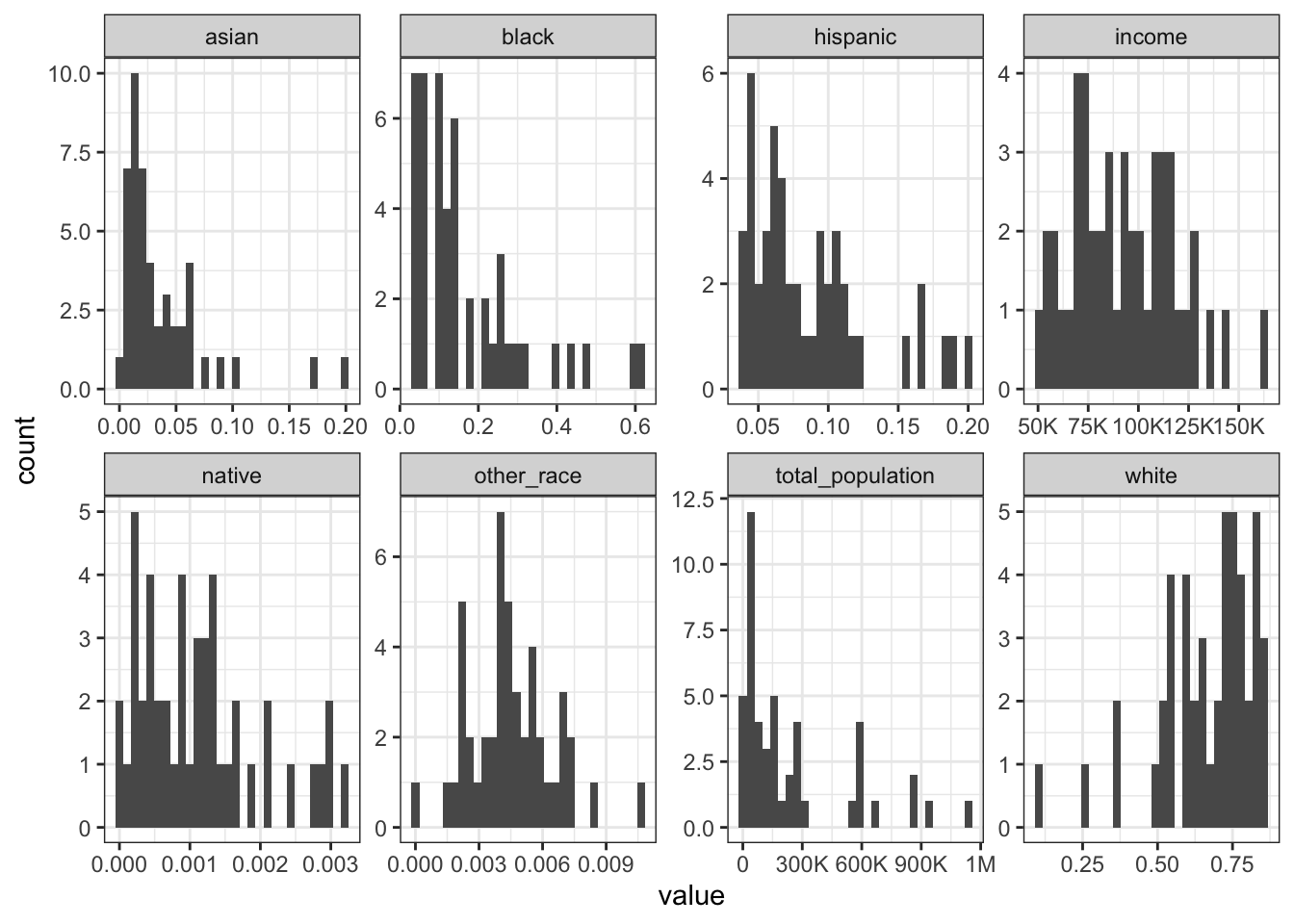
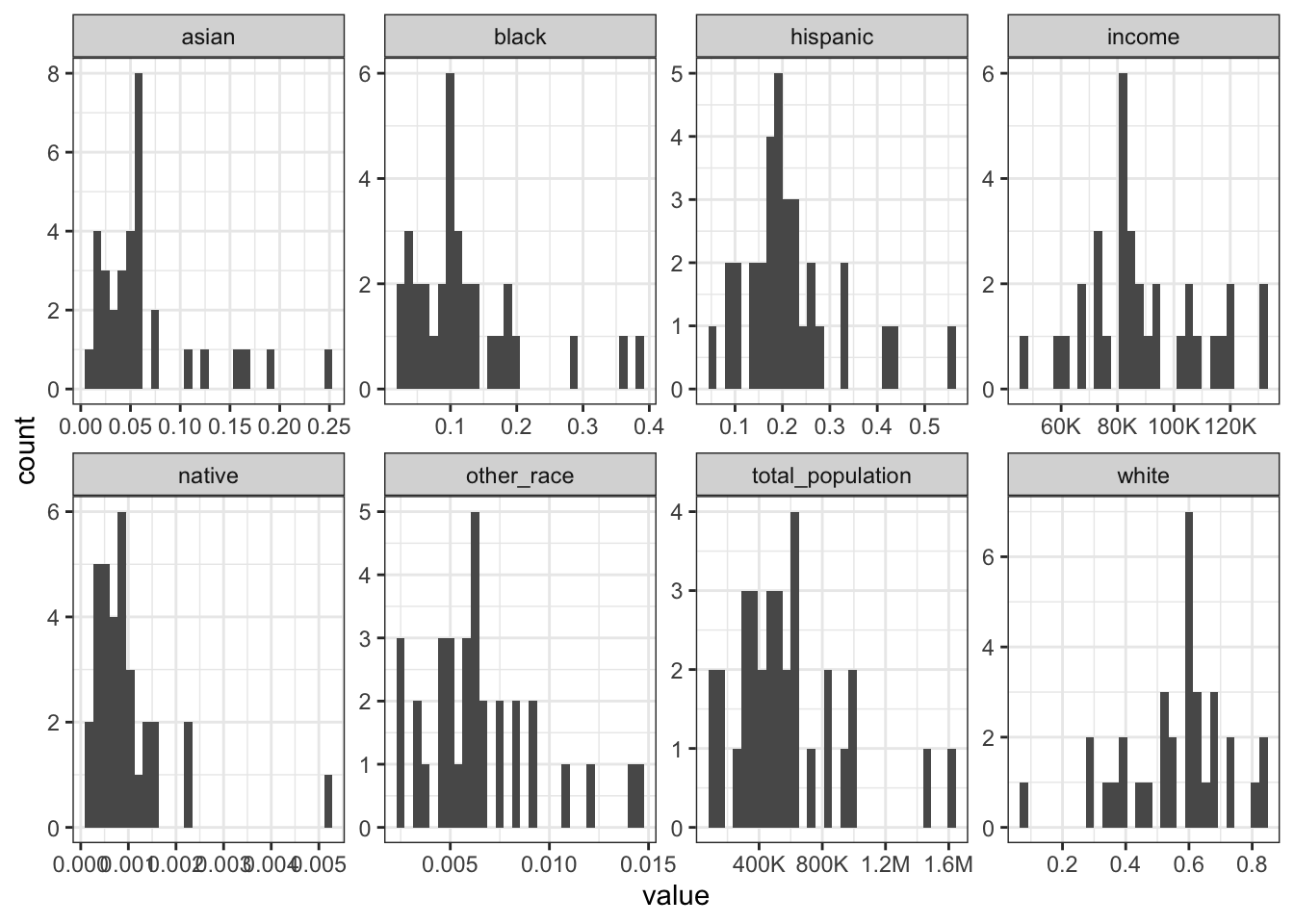
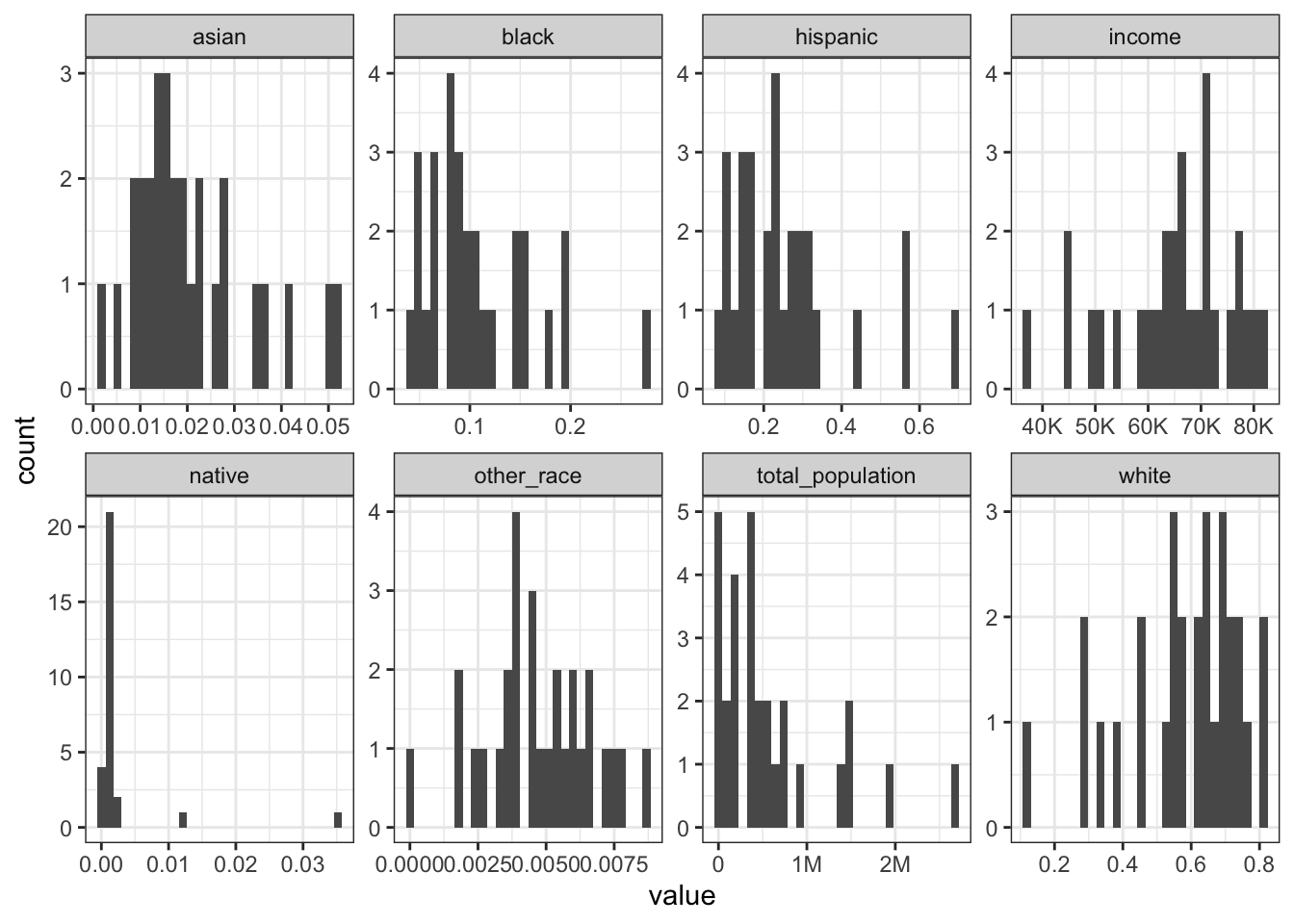
Rows: 1,604
Columns: 17
$ NAME <chr> "Abbeville County, South Carolina", "Accomack Count…
$ geometry <GEOMETRY [°]> POLYGON ((-82.74 34.21, -82..., MULTIPOLYG…
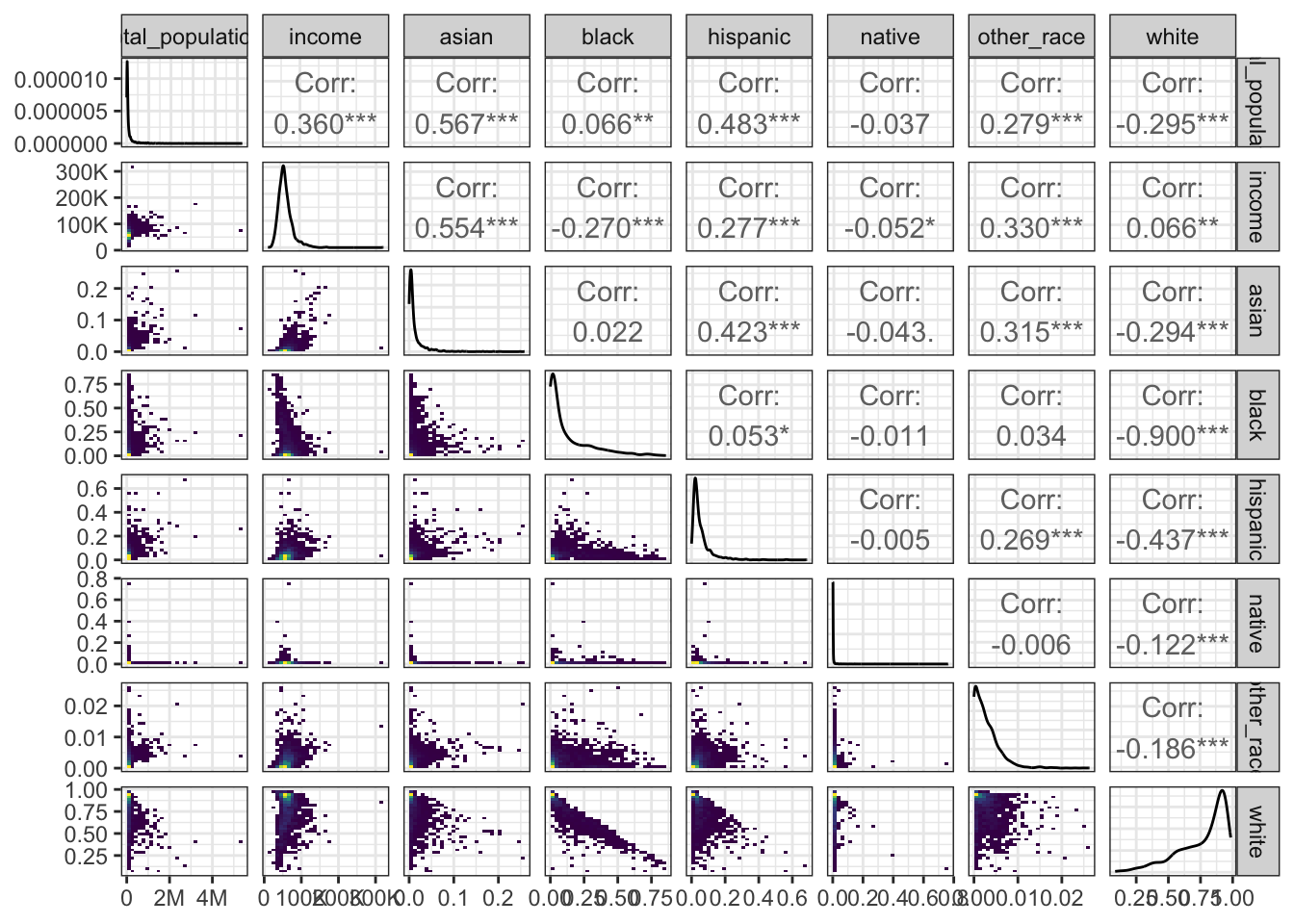
$ dem_asian <dbl> 58, 268, 59, 487, 51, 264, 127, 844, 138, 702, 1696…
$ dem_black <dbl> 6372, 9446, 307, 2565, 213, 15707, 97, 1415, 507, 3…
$ dem_hispanic <dbl> 441, 3084, 468, 1198, 1673, 2051, 303, 7688, 931, 9…
$ dem_native <dbl> 28, 56, 3, 40, 9, 57, 38, 53, 123, 42, 201, 429, 18…
$ dem_other_race <dbl> 138, 55, 6, 96, 39, 158, 36, 328, 118, 55, 500, 140…
$ dem_white <dbl> 16658, 19813, 17308, 59490, 33254, 10834, 26354, 92…
$ income <dbl> 49759, 52694, 49690, 63767, 61731, 37271, 46234, 78…
$ total_population <dbl> 24368, 33367, 18887, 65583, 35827, 29425, 27509, 10…
$ pct_dem_asian <dbl> 0.0023802, 0.0080319, 0.0031238, 0.0074257, 0.00142…
$ pct_dem_black <dbl> 0.261490, 0.283094, 0.016255, 0.039111, 0.005945, 0…
$ pct_dem_hispanic <dbl> 0.018098, 0.092427, 0.024779, 0.018267, 0.046697, 0…
$ pct_dem_native <dbl> 0.0011490, 0.0016783, 0.0001588, 0.0006099, 0.00025…
$ pct_dem_other_race <dbl> 0.0056632, 0.0016483, 0.0003177, 0.0014638, 0.00108…
$ pct_dem_white <dbl> 0.6836, 0.5938, 0.9164, 0.9071, 0.9282, 0.3682, 0.9…
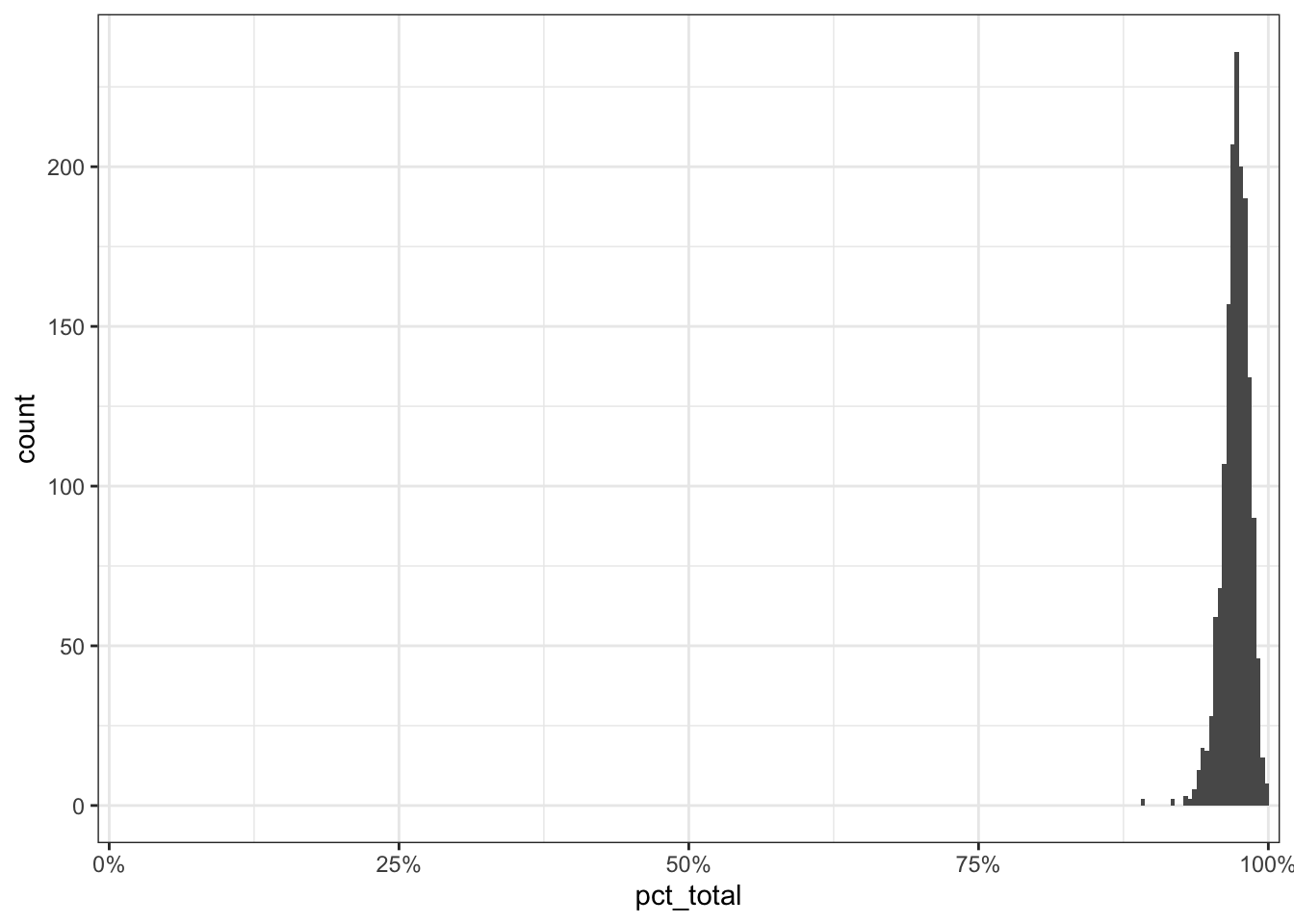
$ pct_total <dbl> 0.9724, 0.9807, 0.9610, 0.9740, 0.9836, 0.9880, 0.9…